julia
using BattMo, GLMakie
model_settings = load_model_settings(; from_default_set = "P2D")
model_settings["SEIModel"] = "Bolay"
cell_parameters = load_cell_parameters(; from_default_set = "Chen2020")
cycling_protocol = load_cycling_protocol(; from_default_set = "CCCV")
simulation_settings = load_simulation_settings(; from_default_set = "P2D")
model_setup = LithiumIonBattery(; model_settings)
cycling_protocol["TotalNumberOfCycles"] = 10
sim = Simulation(model_setup, cell_parameters, cycling_protocol);
output = solve(sim;)
print_output_overview(output)
time_series = get_output_time_series(output)
states = get_output_states(output)
metrics = get_output_metrics(output)(DischargeCapacity = [3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036, 3.6764157469683036 … 3.60571544414199, 3.60571544414199, 3.60571544414199, 3.60571544414199, 3.60571544414199, 3.60571544414199, 3.60571544414199, 3.60571544414199, 3.60571544414199, 0.0], CycleNumber = [0, 0, 0, 0, 0, 0, 0, 0, 0, 0 … 9, 9, 9, 9, 9, 9, 9, 9, 9, 10], RoundTripEfficiency = [77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107, 77.3622897093107 … 87.09682463087034, 87.09682463087034, 87.09682463087034, 87.09682463087034, 87.09682463087034, 87.09682463087034, 87.09682463087034, 87.09682463087034, 87.09682463087034, NaN], ChargeEnergy = [59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256, 59720.469680114256 … 51953.52591462784, 51953.52591462784, 51953.52591462784, 51953.52591462784, 51953.52591462784, 51953.52591462784, 51953.52591462784, 51953.52591462784, 51953.52591462784, 0.0], DischargeEnergy = [45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285, 45308.71046400285 … 44357.379714917864, 44357.379714917864, 44357.379714917864, 44357.379714917864, 44357.379714917864, 44357.379714917864, 44357.379714917864, 44357.379714917864, 44357.379714917864, 0.0], ChargeCapacity = [-4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871, -4.293171612996871 … -3.663400648471446, -3.663400648471446, -3.663400648471446, -3.663400648471446, -3.663400648471446, -3.663400648471446, -3.663400648471446, -3.663400648471446, -3.663400648471446, 0.0])Plot a simple pre-defined dashboard
julia
plot_dashboard(output)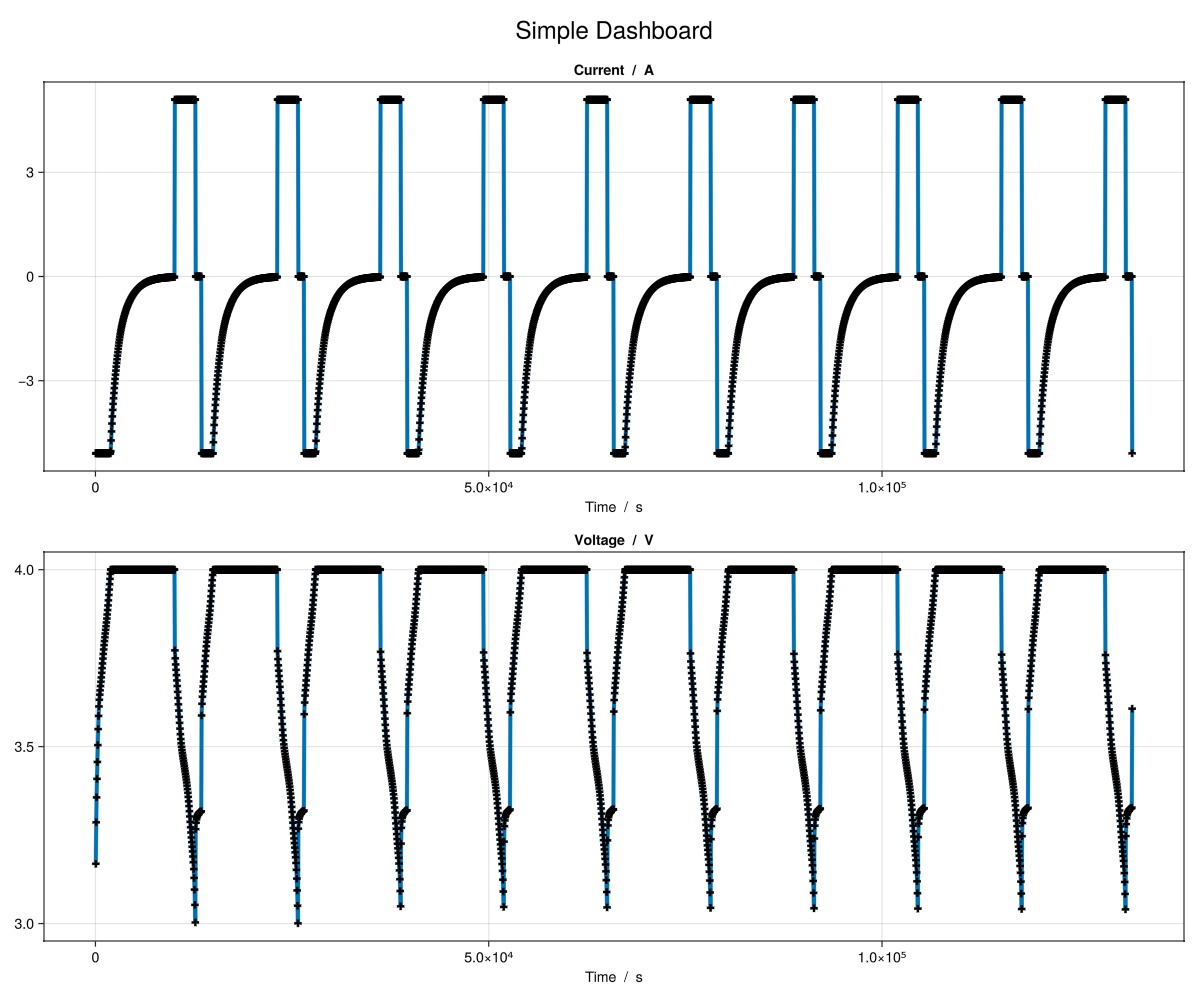
Plot a dashboard with line plots
julia
plot_dashboard(output; plot_type = "line")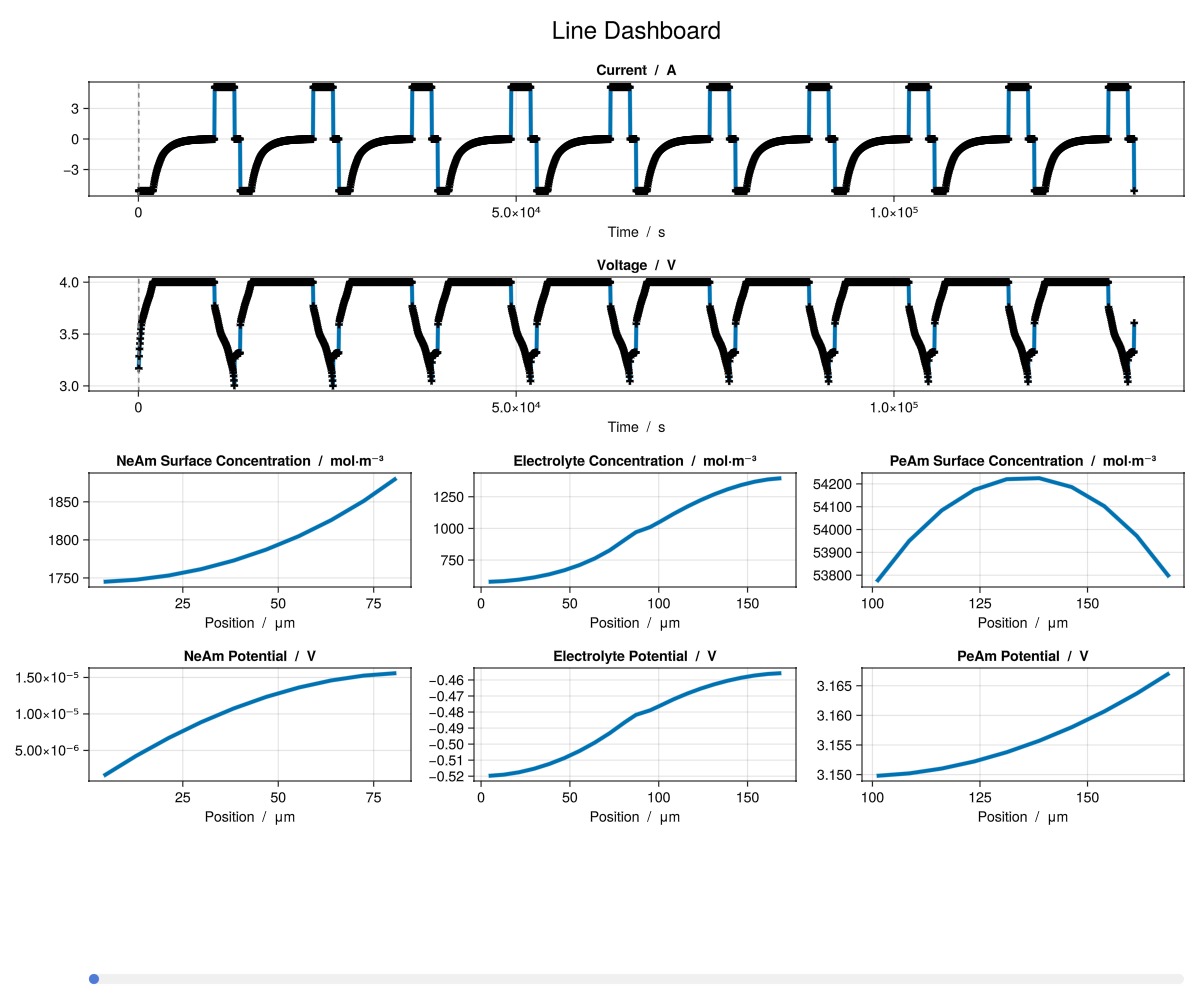
Plot a dashboard with contour plots
julia
plot_dashboard(output; plot_type = "contour")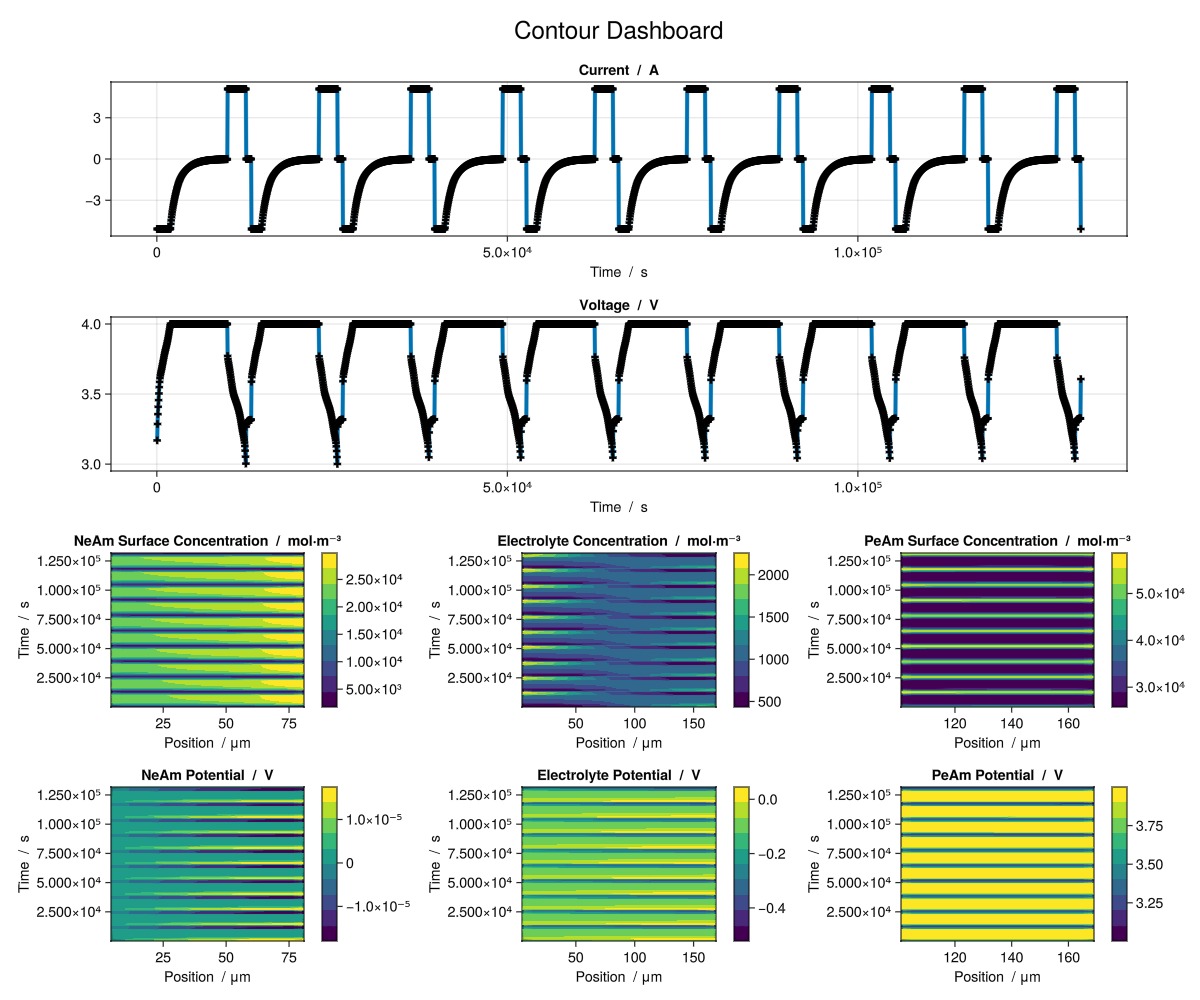
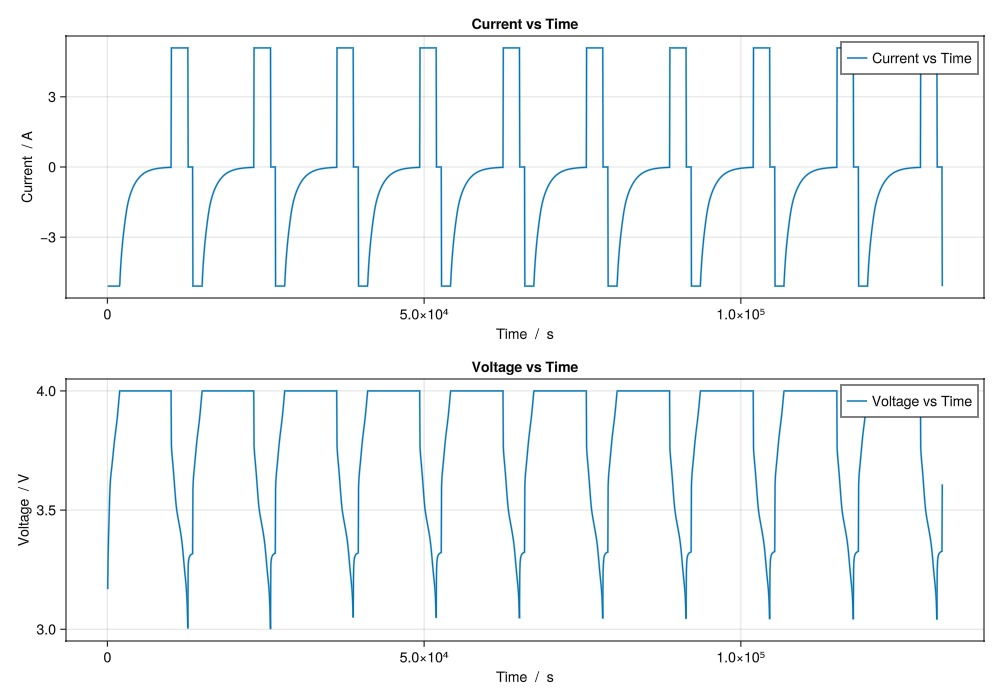
Some simple examples plotting time series quantities using the plot_output function
julia
plot_output(output,
[
"Current vs Time",
"Voltage vs Time",
];
layout = (2, 1),
)
Some simple examples plotting state quantities using the plot_output function
julia
plot_output(output,
[
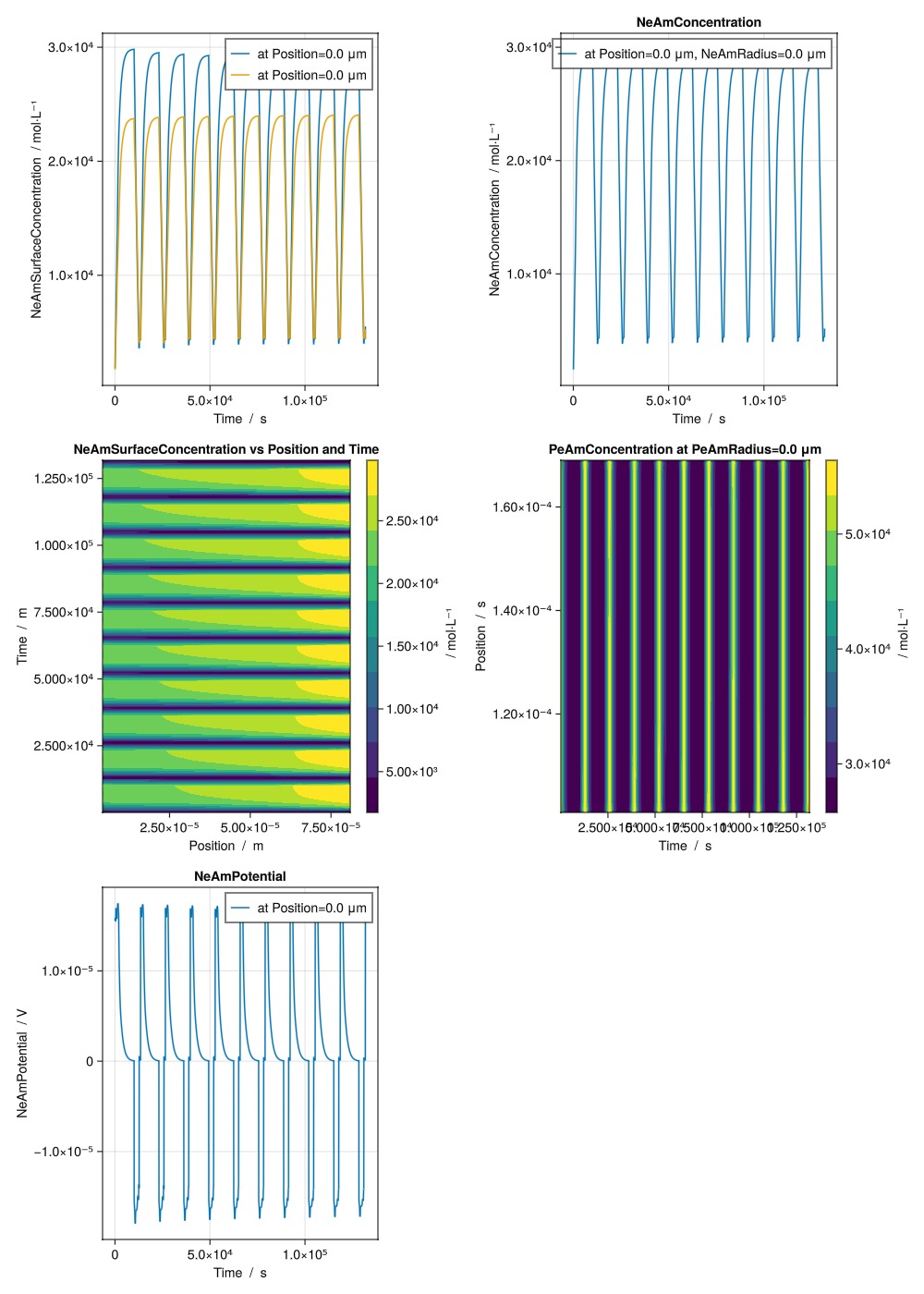
["NeAmSurfaceConcentration vs Time at Position index 10", "NeAmSurfaceConcentration vs Time at Position index 1"],
"NeAmConcentration vs Time at Position index 10 and NeAmRadius index 5",
"NeAmSurfaceConcentration vs Position and Time",
"PeAmConcentration vs Time and Position at PeAmRadius index end",
"NeAmPotential vs Time at Position index 10",
];
layout = (4, 2),
)
Some simple examples plotting metrics using the plot_output function
julia
plot_output(output,
[
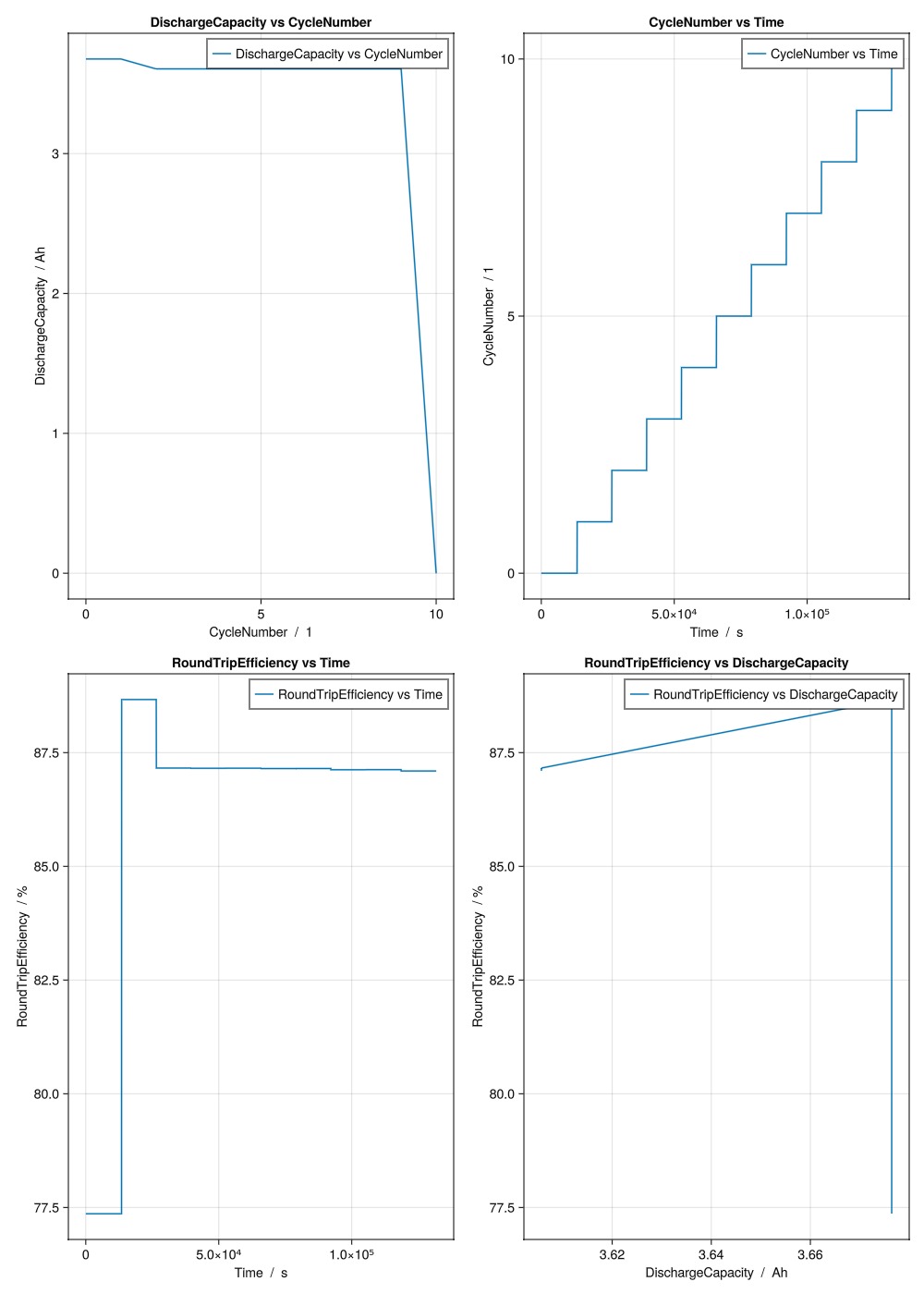
"DischargeCapacity vs CycleNumber",
"CycleNumber vs Time",
"RoundTripEfficiency vs Time",
"RoundTripEfficiency vs DischargeCapacity",
];
layout = (4, 2),
)
Access state data and plot for a specific time step
julia
quantities = ["Time", "Position", "NeAmRadius", "NeAmConcentration",
"NeAmSurfaceConcentration", "PeAmRadius", "PeAmConcentration",
"PeAmSurfaceConcentration", "ElectrolyteConcentration"]
output_data = get_output_states(output, quantities = quantities);
t = 100 # time step to plot
d1 = output_data[:NeAmSurfaceConcentration][t, :]
d2 = output_data[:PeAmSurfaceConcentration][t, :]
d3 = output_data[:ElectrolyteConcentration][t, :]
f = Figure()
ax = Axis(f[1, 1], title = "Concentration at t = $(output_data[:Time][t]) s", xlabel = "Position [m]", ylabel = "Concentration")
l1 = lines!(ax, output_data[:Position], d1, color = :red, linewidth = 2, label = "NeAmSurfaceConcentration")
l2 = lines!(ax, output_data[:Position], d2, color = :blue, linewidth = 2, label = "PeAmSurfaceConcentration")
l3 = lines!(ax, output_data[:Position], d3, color = :green, linewidth = 2, label = "ElectrolyteConcentration")
axislegend(ax)
display(GLMakie.Screen(), f)
g = Figure()
ax2 = Axis(g[1, 1], title = "Active Material Concentration at t = $(output_data[:Time][t]) s", xlabel = "Position", ylabel = "Depth")
hm1 = contourf!(ax2, output_data[:Position], output_data[:NeAmRadius], output_data[:NeAmConcentration][t, :, :])
hm2 = contourf!(ax2, output_data[:Position], output_data[:PeAmRadius], output_data[:PeAmConcentration][t, :, :])
Colorbar(g[1, 2], hm1)
display(GLMakie.Screen(), g)GLMakie.Screen(...)Example on GitHub
If you would like to run this example yourself, it can be downloaded from the BattMo.jl GitHub repository as a script.
This page was generated using Literate.jl.